
Introduction: Additional Sex Combs-Like 1 (ASXL1) is a chromatin modifier frequently affected by truncating mutations in myeloid malignancies. In chronic myelomonocytic leukemia (CMML), truncating ASXL1 mutations are associated with the overexpression of leukemogenic driver genes, however the molecular mechanisms underlying this transcriptional activation remain controversial. We performed single-cell chromatin accessibility studies of ASXL1-mutant (MT) and -wildtype (WT) CMML to identify cis-regulatory elements (CREs) such as distal enhancer elements that may explain the observed transcriptional activity in MT CMML.
Methods: Bone marrow mononuclear cells from a patient with WT and a patient with MT CMML were subjected to single-cell DNA transposase accessibility studies (scATAC-seq). The cryopreserved cells were thawed and resuspended and approximately 100000 viable mononuclear cells per sample were subjected to transposase assays before proceeding to single-cell partitioning into gel beads in emulsion, barcoding, library construction, and sequencing following an established 10X Genomics workflow. The target cell recovery was approximately 2000 cells per sample. Genomic libraries were sequenced on an Illumina HiSeq 4000 before demultiplexing, alignment to the reference genome, and post-alignment quality control. The 10X Genomics Cell Ranger ATAC software was used for demultiplexing, alignment of the reads to the GRCh38 reference genome, filtering and quality control, counting of barcodes and unique molecular identifiers, identification of transposase cut sites, detection of accessible chromatin peaks, count matrix generation for peaks and transcription factors, clustering, and differential accessibility analysis. The two-sample Kolmogorov-Smirnov test for equality of distribution functions was used to compare the distributions of single-cell entropies. Single-cell cis-co-accessibility scores were estimated using Cicero.
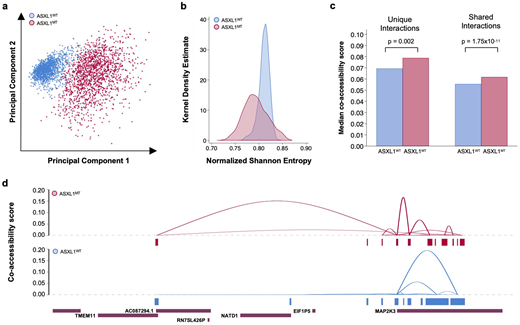
Results: To mitigate potential effects on the analysis introduced by unequal cell recovery due to experimental conditions, we randomly sampled 1500 single cells from each genotype. Visualizing the first two principal components of the chromatin accessibility data revealed the differences in chromatin conformation between genotypes and also the increased heterogeneity within the MT sample (Figure 1a). To quantify this increase in intratumoral heterogeneity of chromatin accessibility in MT CMML, we calculated the single-cell entropies for both genotypes. There was an increase in the dispersion of single-cell entropy values (mean 0.793±0.026 versus 0.811±0.012, p=6.19x10-151) in MT compared to WT disease (Figure 1b). To leverage the power of the single-cell chromatin accessibility data, we estimated cis-co-accessibility scores (two-way chromatin interactions). There were 631 genes involved in two-way chromatin interactions that were either specific to MT CMML or were shared between MT and WT CMML and showed an increase in co-accessibility in MT CMML. Co-accessibility was higher in MT compared to WT CMML, regardless whether the chromatin interaction was MT-specific or shared between the genotypes (Figure 1c). Gene ontology analysis revealed increased chromatin accessibility in genes involved in the regulation of cell cycle, nuclear division, chromosome organization, cell cycle phase transition, DNA conformation change, and DNA replication (FDR<1.00x10-5). The construction of cis-co-accessibility networks demonstrated a remodeling of the chromatin landscape with MT-specific chromatin interactions involving key mitotic kinases such as MAP2K3 (Figure 1d).
Conclusions: MT CMML is characterized by an increase in intratumoral heterogeneity and increased chromatin accessibility. The increased chromatin co-accessibility involved key mitotic kinases and may serve as a plausible explanation for the increased transcriptional activity observed in MT CMML.
Ordog:Millipore Sigma: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This icon denotes a clinically relevant abstract

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal